This article was also written by Dr. Azam Ali Sher.
Campylobacter, a pathogenic bacteria often linked with poultry, is a leading cause of food poisoning in the United States. The US Centers for Disease Control and Prevention (CDC) estimates that each year, there are 1.5 million human cases of campylobacteriosis in the US, which generate approximately $270 million in direct medical costs.
C. jejuni’s Impact
C. jejuni, a specific strain of Campylobacter, isn’t just the leading cause of food poisoning nationwide. It also disproportionately affects US military personnel and can cause a rare neurological disorder.
Campylobacter can survive in many places including the gastrointestinal tracts of humans and many animals, as well as environmental sites like farms or other food production environments. Pathogens like Campylobacter often evolve to become better-able to survive in new environments. And like many other pathogens, Campylobacter is affected by the growing problem of antibiotic resistance. A recent CDC report found that 29 percent of Campylobacter infections in the US were drug-resistant. Antibiotic-resistant C. jejuni infections can last longer, be more difficult to treat, and cause more serious illness.
Without insight into the mechanisms of a specific bacterium’s evolution, it’s more difficult to understand how antibiotic resistance emerges and, similarly, to find new therapeutic and preventative treatment options—in this case, for C. jejuni-based food poisoning. But that is what one Spartan research team is working to understand.
The Study
What is the Growth Medium, and How Did They Use it?

The team used Bolton Broth, a nutrient-enriched growth medium, to study the evolution of C. jejuni. Bolton Broth contains nutrients that help facilitate the growth and development of C. jejuni while offering the pathogen protection from oxygen and other harmful exposures. Prior to this study, Bolton Broth had not yet been used to study these specific C. jejuni populations.
Past studies of Campylobacter’s evolution have used laboratory-adapted bacterial strains, rather than fully pathogenic, clinically relevant strains like Campylobacter jejuni, which is the species responsible for most foodborne illness in humans. This is important because laboratory-adapted strains and clinically relevant strains evolve differently because they exist in different environments. (For example, laboratory-adapted strains often cannot propel themselves on their own, due to evolving over several generations in the cushy environment of growth medium. However, that ability to move with external assistance, known as motility, is essential for infecting a host naturally.)
To understand how host-adapted, motile C. jejuni evolves to grow and thrive, the research team set up an experiment that featured the novel use of a controlled environment: nutrient-enriched laboratory growth medium.
In the study, C. jejuni bacteria that were motile were passaged and isolated from the gut microbiomes of mice. Only fully motile bacteria remained after this process. Later, these different bacterial populations were grown in the lab using Bolton Broth. During this phase, the scientists observed the bacteria’s loss of motility and other associated genomic changes.
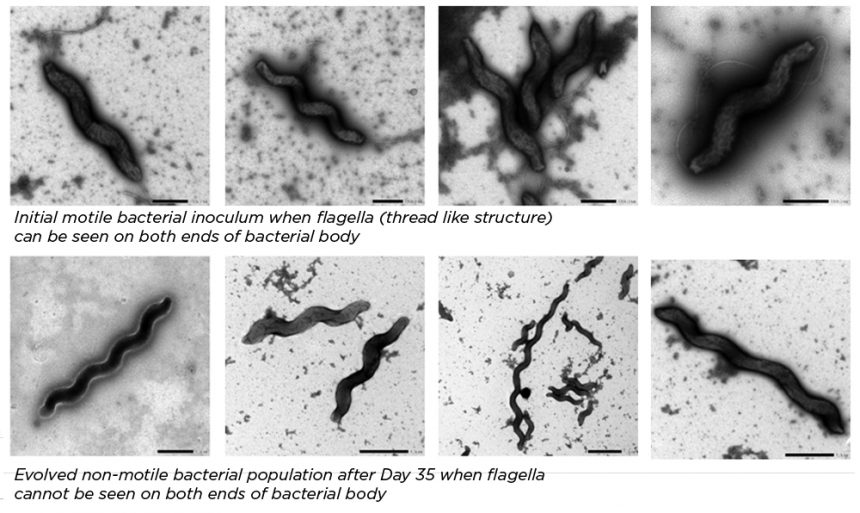
What is a flagellum, and why does it matter?
The flagellum is the appendage that allows a bacterium to move, or to be motile. In this experiment, an electron microscope, which provides very high-resolution images, revealed a total loss of flagella in one evolved population, compared to the initial inoculum of bacteria introduced to the medium.
“This project design allowed us to compare how C. jejuni evolves in the artificial, nutrient-rich environment of growth medium, where everything it needs to survive is automatically available and accessible,” says Dr. Azam Ali Sher, first author on the study published by the research team. “This is different from the natural mammalian gut, where C. jejuni must move itself around to colonize and survive.”
After the research team subcultured five mouse gut-adapted populations of C. jejuni into fresh growth medium, they continued their study; for 35 days, they transferred 100 microliters of subcultures to the fresh medium every 24 hours. Throughout these 35 days, the team observed changes in the motility of the bacterium and noted three types of motility across the five C. jejuni populations:
- Reversibly non-motile (cannot move itself, but this could change)
- Irreversibly non-motile (cannot move itself, and this could not change)
- Motile (can move itself)
After the first 5 days, a significant portion of the C. jejuni colonies were non-motile. By day 20, the irreversibly non-motile form dominated. This loss of motility continued to increase. By day 35, approximately 80 percent of all 5 populations were non-motile.
Digging into the DNA
What led to this motility loss? To find out, the team performed whole genome re-sequencing on the five evolved populations of C. jejuni. This means they looked at the bacteria’s entire genome, or the entire set of genetic material, to reveal each population’s genetic “fingerprint.”
The team detected many disruptive mutations in the genes of the C. jejuni populations associated with the flagellum. Of note, they observed that the gene rpoN had a high number of deletions and also contained multiple pseudogenes. Testing revealed that bacteria with changed, or mutated, rpoN genes grew markedly faster in the growth medium than the initial strain of C. jejuni. Essentially, the non-motile bacteria grew faster in the broth than the motile bacteria did.
About the researchers
Dr. Azam Ali Sher, PhD student at the MSU College of Veterinary Medicine, works in the laboratory of Dr. Linda Mansfield, University Distinguished Professor and Albert C. and Lois. E. Dehn Endowed Chair in Veterinary Medicine. Sher is first author on the team’s latest paper published in Frontiers in Microbiology.
Craving more of the science? Check out Dr. Azam Ali Sher’s article in Frontiers Science News for more discussion of the research
“This work indicates that C. jejuni undergoes genome reduction when grown in rich media and suggests that its evolutionary mechanisms in different hosts and environmental niches should be explored,” says Mansfield.
Looking Ahead: Therapeutic and Preventative Possibilities
While further study is required to confirm which mutations triggered the motility loss, and Sher notes that in vitro studies should be interpreted cautiously, the team’s findings could prove useful in the future for patients suffering from campylobacteriosis.
“It is unlikely that motility loss could increase the virulence or survival of the bacteria in a host,” explains Sher. “But experimental evolution studies like ours targeting other genes needed to cause disease could be an effective tool to discover novel drug targets and vaccine candidates against pathogenic bacteria like C. jejuni.”
To learn more about this research, contact Dr. Linda Mansfield.
